Anisotropic Network Model¶
Files for this example can be downloaded from here.
This example is located in mdtoolbox_example/anm_lys/.
Normal mode analysis of ANM¶
Normal mode analysis of Ca-based anisotropic network model of T4 lysozyme (script_anm.m).
%% Nomal mode analysis of anisotropic network model of T4 lysozyme
% read Ca coordinates from PDB file
[pdb, crd] = readpdb('lys.pdb');
index_ca = selectname(pdb.name, 'CA');
crd = crd(to3(index_ca));
crd = decenter(crd);
% normal mode of anisotropic network model (ANM)
[emode, frequency, covar, covar_atom] = anm(crd, 8.0);
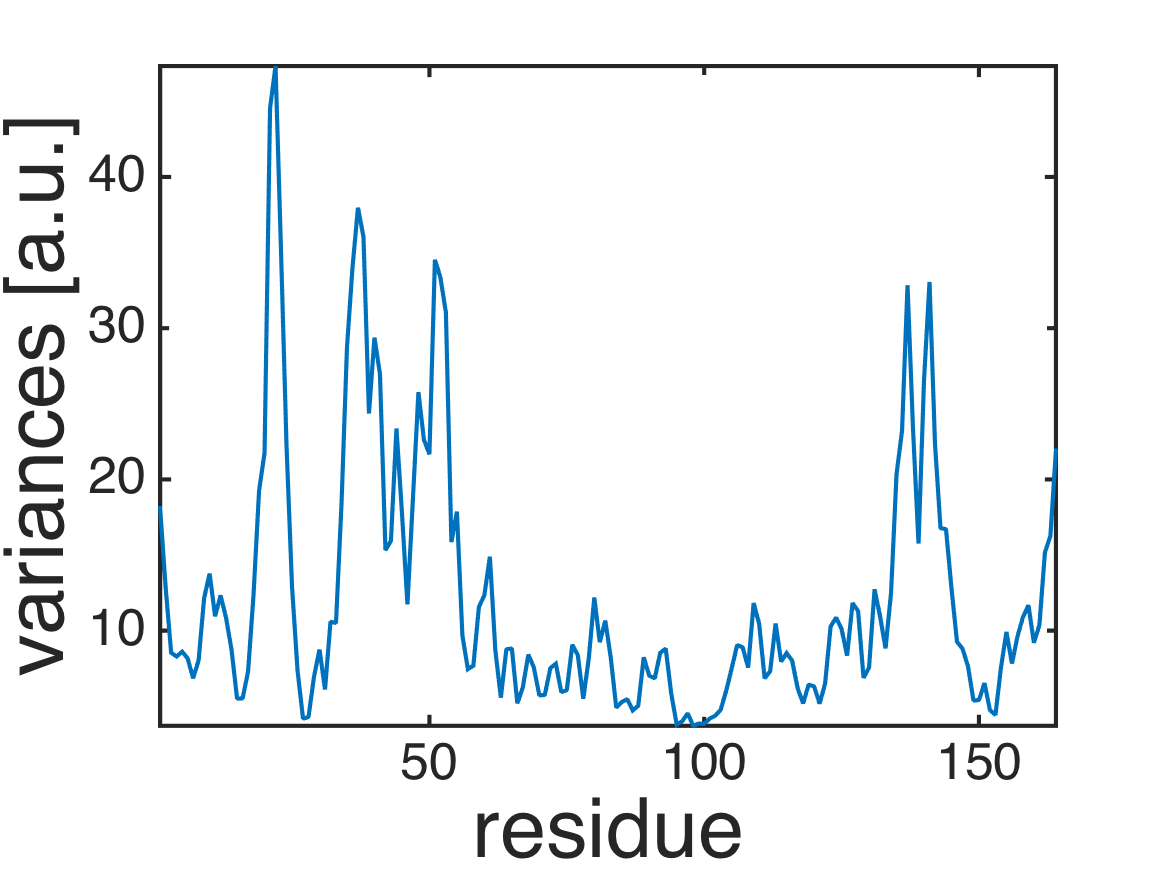
% plot root-mean-square-fluctuations (RMSF)
plot(diag(covar_atom));
axis tight;
xlabel('residue','fontsize',40);
ylabel('variances [a.u.]','fontsize',40);
formatplot
exportas('rmsf');
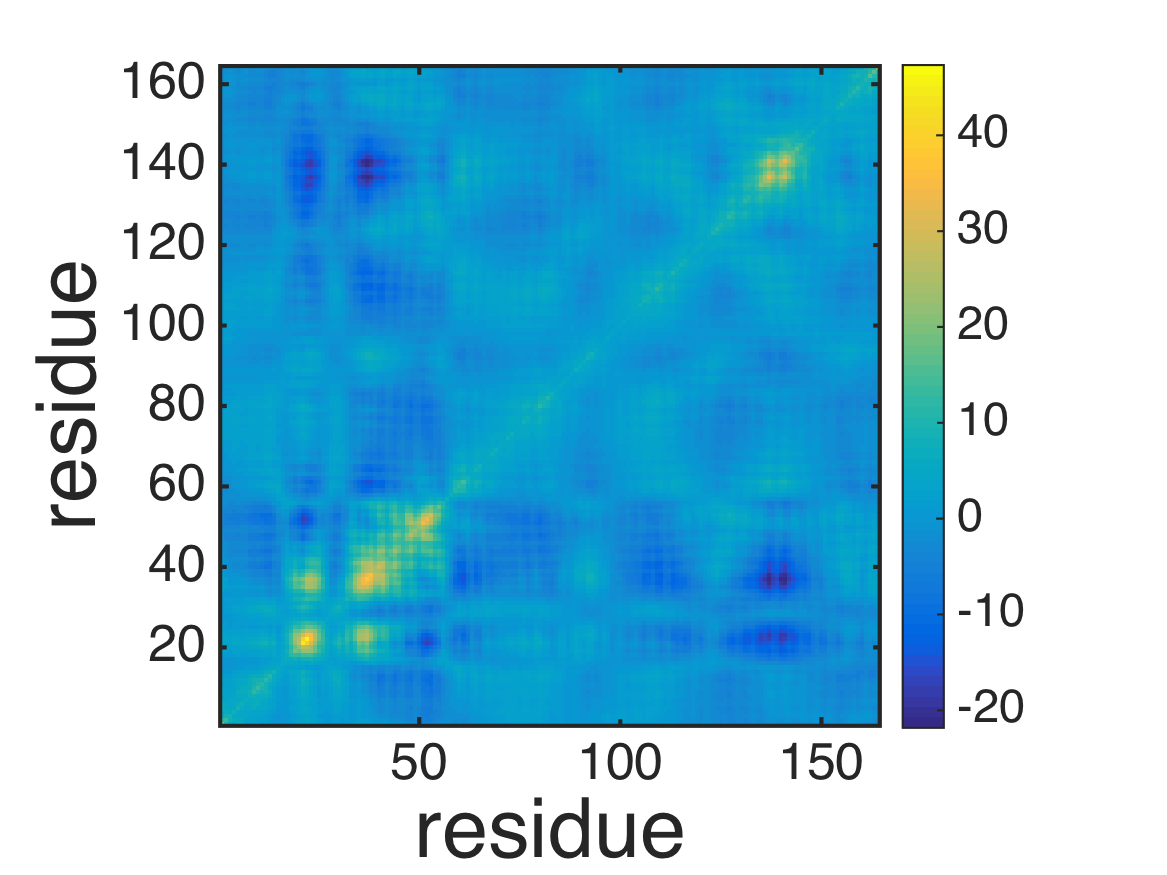
% plot covariance
imagesc(covar_atom);
axis xy; axis square;
xlabel('residue','fontsize',40);
ylabel('residue','fontsize',40);
colorbar;
formatplot2
exportas('covar_atom');
% export NMD file for visalizing mode structures
writenmd('anm.nmd', crd, emode);
% save data
save script_anm.mat;
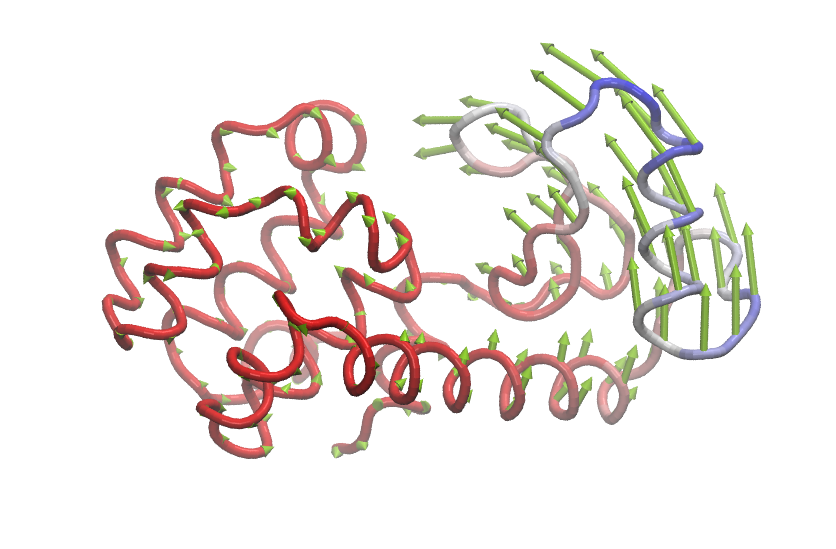
Visualize mode structures by using the Normal mode wizard in VMD.
$ vmd
vmd > nmwiz load anm.nmd
Transformation of frame¶
Transform from the Eckart frame to a non-Eckart frame (script_transformframe.m).
%% Transform from the Eckart frame to a non-Eckart frame.
% load data
load script_anm.mat;
% transform frame
index_fixeddomain = [1:11 77:164]; %atom-index for the larger domain
external_mode = emode(:,(end-5):end);
[emode2, variances2, covar2, covar2_atom] = transformframe(index_fixeddomain, external_mode, covar);
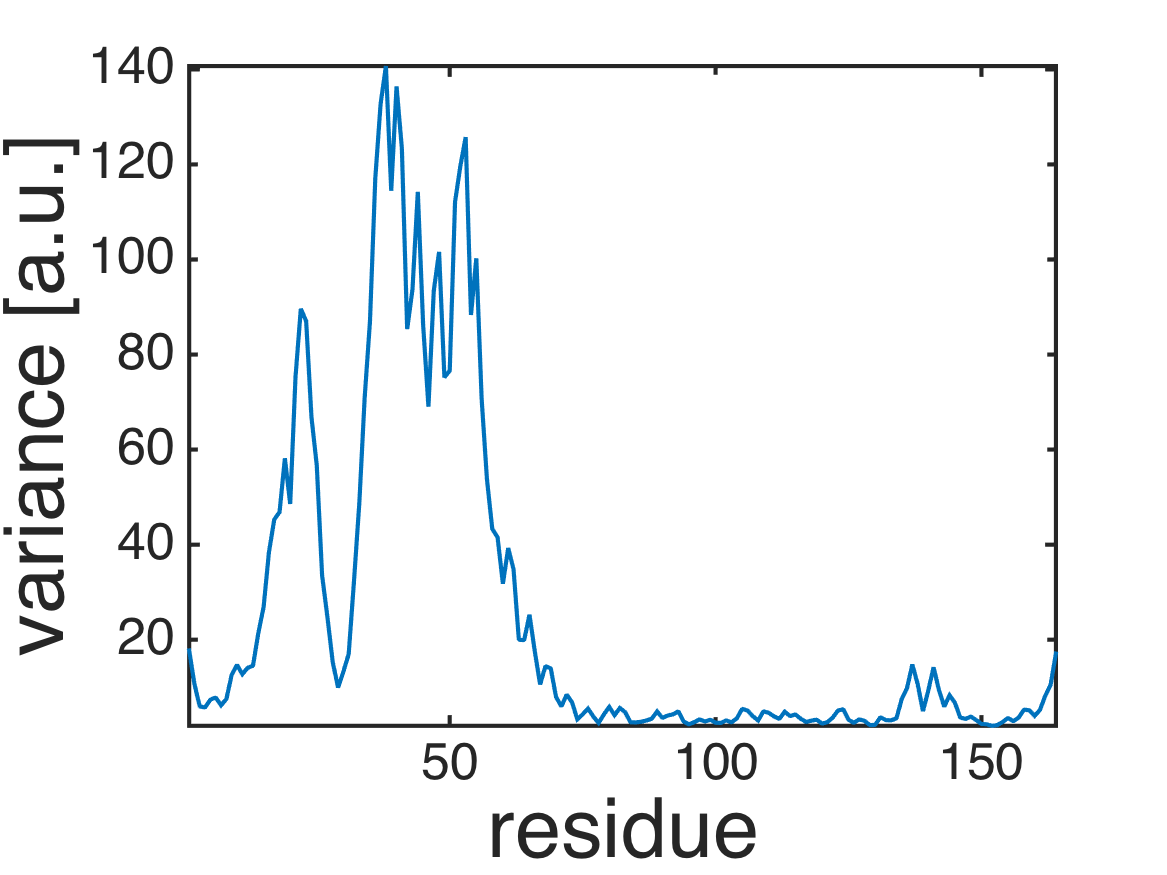
% plot root-mean-square-fluctuations (RMSF)
plot(diag(covar2_atom));
axis tight;
xlabel('residue','FontSize',40);
ylabel('variance [a.u.]','FontSize',40);
formatplot
exportas('rmsf_ne');
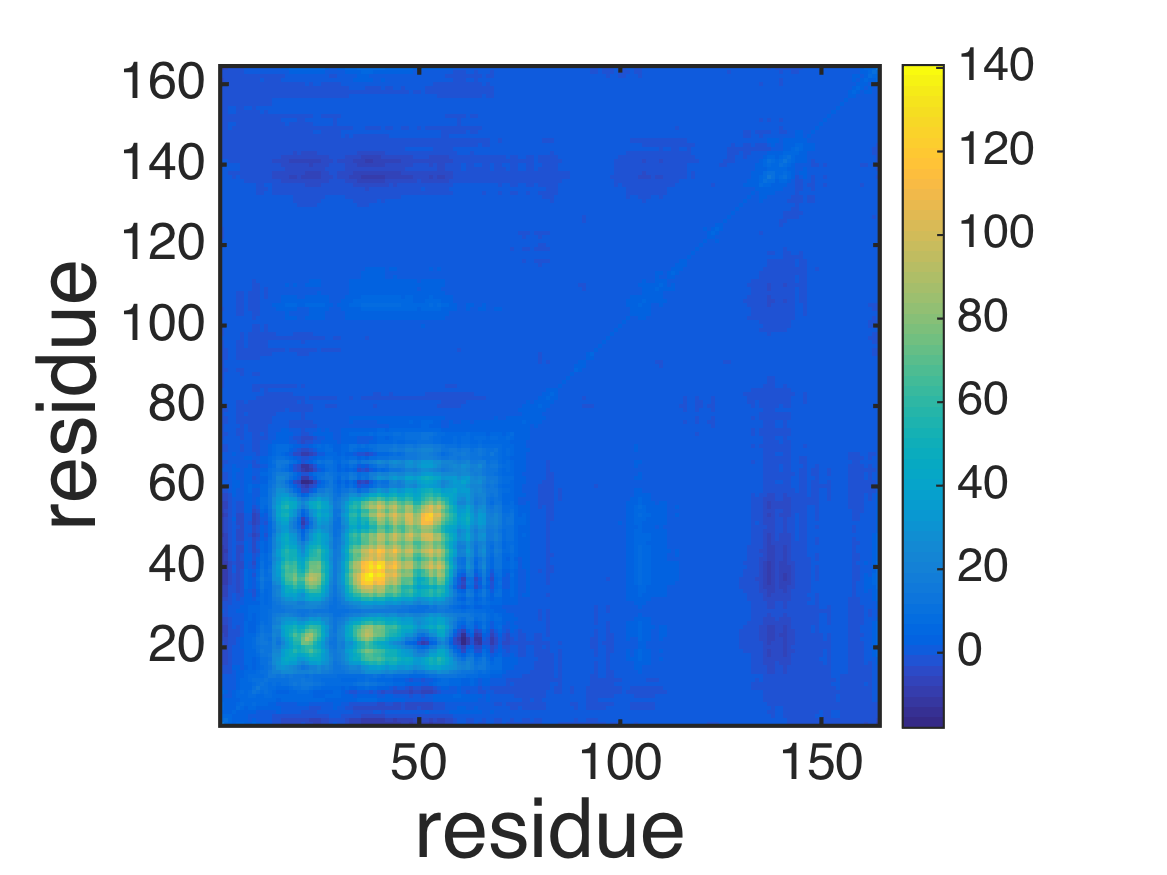
% plot covariance
imagesc(covar2_atom);
axis xy; axis square;
xlabel('residue','FontSize',40);
ylabel('residue','FontSize',40);
colorbar;
formatplot2;
exportas('covar_atom_ne');
% export PDB files for visalizing mode structures
writenmd('anm_ne.nmd', crd, emode2);
% save data
save script_transformframe.mat;
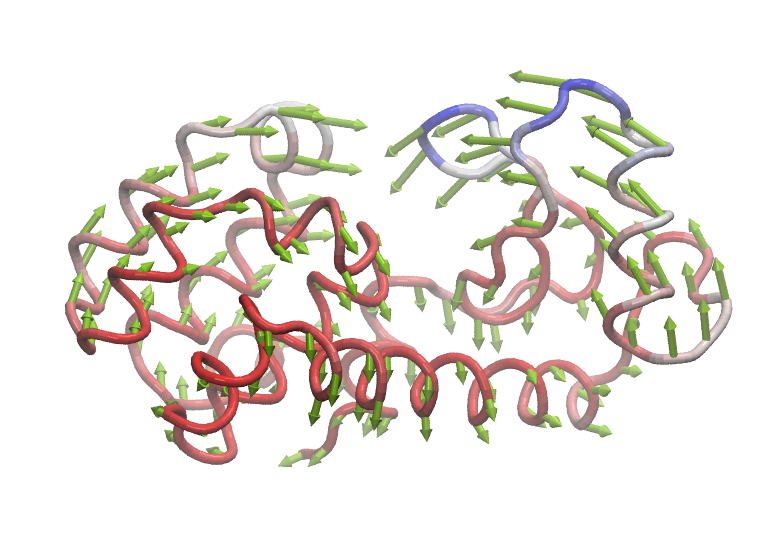
Visualize mode structures by using the Normal mode wizard in VMD.
$ vmd
vmd > nmwiz load anm_ne.nmd